It brings them closer to their goal of being able to engineer the assembly of DNA brick structures so that they could be used in applications such as diagnostics. And there are other potential applications in the fabrication of devices, such as nanoscale logic circuits and data structures, that could be the building blocks of a future 'nano computer'.
Researchers here in the department, in collaboration with scientists at the Fraunhofer Institute in Leipzig, have been studying microscopic self-assembling molecular structures for some time in order to advance our understanding of the physics behind them. Their latest work has focused on DNA brick construction techniques and how they can be used to 'print' precise arrangements of molecules onto surfaces, for example to create large arrays that could be used in some diagnostic applications.
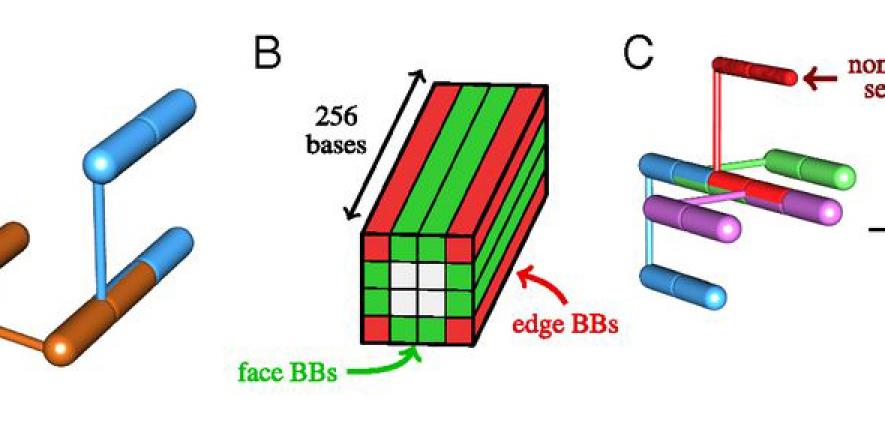
The researchers knew that when dealing with systems involving DNA brick structures, problems can occur. For example, it can take several days for the structures to assemble. And the yield of correctly former structures at the end of the process can often be very low – less than one per cent. These structures are formed in systems that have thousands of distinct components and if just one molecule bonds in the wrong place, and other molecules mistakenly bond with it, it can then frustrate the growth of the target structure.
But in a paper just published in PNAS (Proceedings of the National Academy of Sciences), the Cambridge-Leipzig team demonstrates a simple strategy for optimising the nucleation, or initial, step in the self-assembly of a multicomponent DNA brick structure. As a result, they are also able to offer some protocols for the assembly process, in order to improve the yields of structures created, from less than one per cent up to 25 per cent.
In order to understand the process, so that they could try to optimise it, the researchers used the technique of dynamic light scattering for the first time in the context of DNA self-assembly to probe the assembly process.
"It worked very well, allowing us to observe the process in real time," says Dr Aleks Reinhardt, a researcher here. "Other approaches, like fluorescence measurements, require us to stop the assembly process and also, the fluorescent dyes used can themselves interfere with the process. DLS is less invasive and also much more sensitive."
"Combining that with a theoretical model helped us to show that the ultimate yield of correctly-assembled structures is largely determined by the nucleation pathway. And that allowed us to establish some design principles for improving the self-assembly of addressable nanoscale structures."
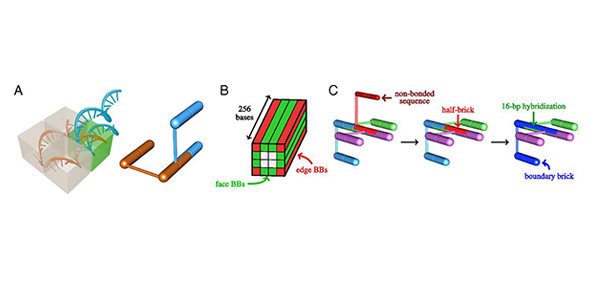
All of this work follows on from the initial discovery, made by scientists at Harvard in 2012, that potentially thousands of distinct DNA molecules can reproducibly self-assemble into complex, fully addressable and nearly error-free target structures.
Making the structures 'addressable' – i.e. where each component is unique and in an exactly-specified position – is vitally important for constructing nanoscale devices as, by controlling their structure, scientists can control their function.
"The 2012 discovery came as something of a shock to the self-assembly community," Aleks explains. "This was not meant to happen, as it had been well-established that having such a large number of distinct components would lead to self-poisoning and in turn a kinetic arrest of the growth of the target structure.
"I thought the findings were really interesting, so I started researching how structures like these can form successfully whilst other systems have failed to achieve anything even approaching such complexity."
Attending a DNA conference, Aleks met scientists from the Fraunhofer Institute who were also studying self-assembling DNA structures. When they combined Aleks' theoretical modelling work with their experimental research, it led over time to an understanding of how nucleation – the initial step in the construction process – plays a very significant role in successful DNA brick structure assembly and how it can be optimised.
As the researchers say in their paper: "In a typical annealing protocol, structures have a limited time and temperature window in which to form: at high temperatures, a large free-energy barrier inhibits nucleation, while at low temperatures, self-assembly is limited by kinetic arrest. The key to successful self-assembly is to increase the width of the temperature window over which nucleation can occur, thereby maximizing the thermodynamic segregation between the critical nucleation step and detrimental misbonding."
One result of being able to optimise the assembly process is that it can now be speeded up by a factor of ten, allowing structures to be assembled in a day rather than a week.
"We’re hope that this will stimulate experimental efforts towards developing practical applications," Aleks says.
- Read the paper here on Direct observation and rational design of nucleation behavior in addressable self-assembly