
Professor of Biophysical Chemistry
Research in TheLeeLab centers on developing new biophysical methods to answer fundamental biological questions, primarily through single-molecule fluorescence spectroscopy and multidimensional super-resolution imaging. Professor Lee completed his DPhil in Physical Chemistry with Dr. Mark Osborne, before postdoctoral work with both Prof Sir David Klenerman (FRS MedSci) and Prof W.E Moerner (Nobel Prize Chemistry 2014). He manages a research team of talented researchers who profoundly believe molecules should be looked at one at a time. Professor Lee is the 2017 recipient of the Marlow Prize in Physical Chemistry from the Royal Society of Chemistry.
Prof Lee also co-hosts a scientific communication podcast, TheScienceShed, and can be contacted via twitter @SteveTheChemist

Research Interests
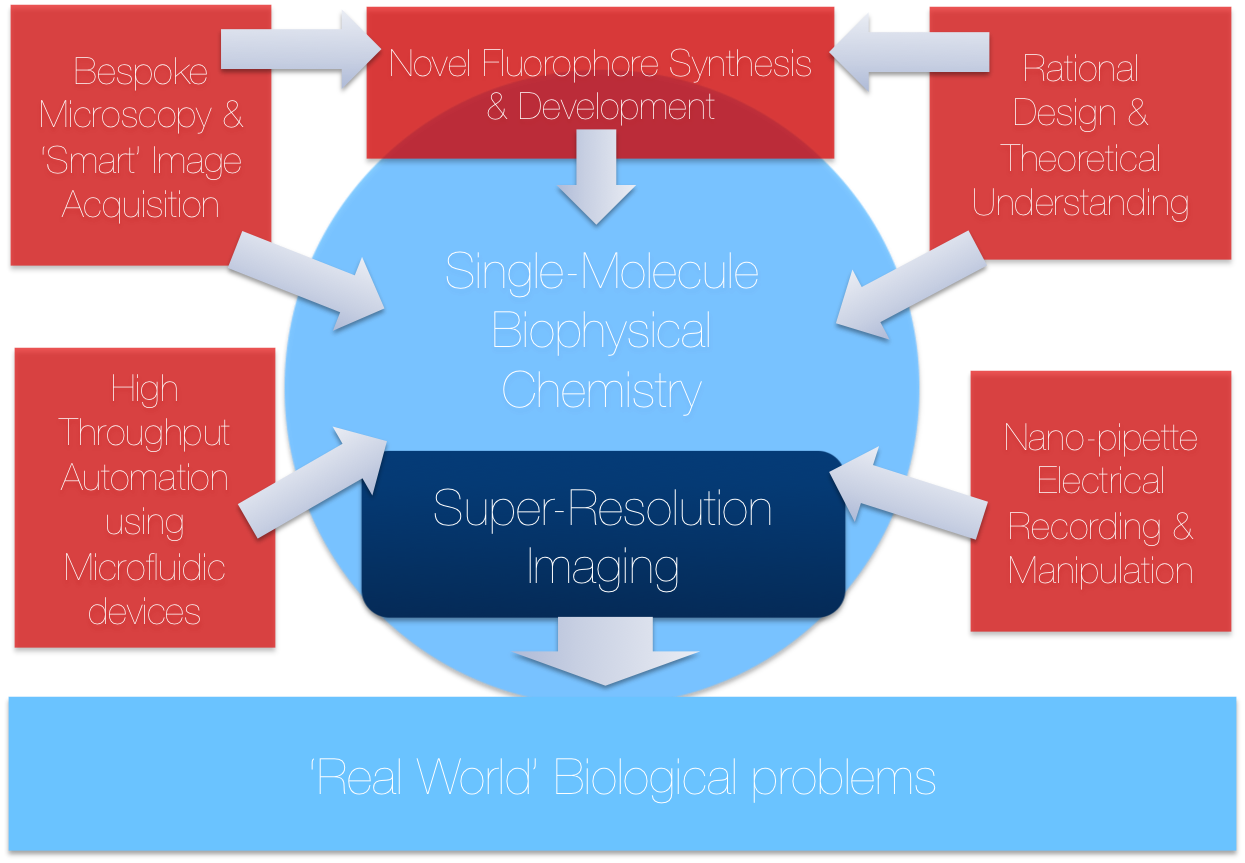
Located in the Chemical Laboratories at the University of Cambridge, TheLeeLab focusses on the development of new biophysical and single-molecule fluorescence methods to answer fundamental biological problems. TheLeeLab focuses on many problems related to new biophysical method development, some of which are detailed below:
Professor Lee discusses his research
Take a tour of the Lee lab
Background
There is a fundamental limit to how small we can see objects, caused by the diffraction of light itself, this is called Abbe's diffraction limit. This innate 'blurriness' means that optical microscopy cannot resolve anything smaller than ~250 nanometres. Unfortunately the spatial scale that many biological processes occur on is smaller than this limit, therefore it has been difficult to directly probe these events.
Super-resolution microscopy is an advanced, interdisciplinary optical imaging technique currently attracting immense interest as a new and exciting way to break this theoretical diffraction limit of visible light. Using single molecule control, super-resolution microscopy retains the non-invasive advantages of fluorescence imaging but has the ability to resolve biological structures more compatible with the spatial scale that these events actually take place on, typically attaining resolutions of ~15nm or better.
We use novel approaches of super-resolution microscopy in both 2D and 3D, to directly visualise biological processes going on inside living cells at two physical areas: in the eukaryotic plasma membrane and in the nucleus, highlighted by two key areas:
1) The molecular origins of human immunity by studying the spatial distribution and interaction of individual membrane protein complexes on the plasma membrane of live T-cells during triggering.
2) How the molecular composition, stoichiometry and spatial arrangement of histones assemble during DNA replication and repair, all inside the nucleus of fission yeast in three dimensions.
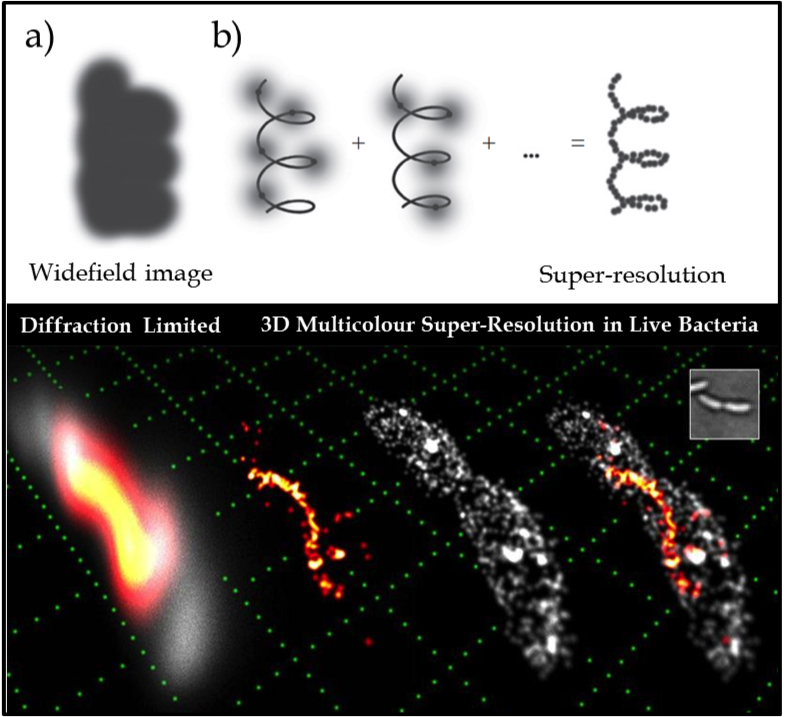
Figure 1. (Upper) The key idea of super-resolution imaging of a structure. (a) It is not possible to resolve the underlying structure in a conventional widefield fluorescence image because the fluorescent labels are in high concentration and the individual emitters overlap. (b) Using controllable fluorophores, it is possible to ‘turn on’ and image a sparse subset of molecules which can then be localised with nanometre precision (grey line is the underlying structure being sampled). Once the first subset of molecules photobleaches, another subset is turned on and localised. This process is repeated and the resulting localisations are summed to give a super-resolution image of the underlying structure. (Lower) Example of 3D two colour SR imaging of the helical cytoskeletal protein crescentin in the live bacterium caulobacter crescentus (shown in orange/red) in relative context to the cellular membrane (grey) attaining lateral and axial precisions of 15 and 20nm respectively. Conventional diffraction limited imaging is shown (far left) as a reference, compared with the exploded view of the two component channels (middle images) and then combined (far right). A white light image (inset) and a reference grid (green) made up of 1µm squares is shown for scale.
These imaging strategies can be applied to a host of other problems where there is need for watching key biological process unfold at high spatial precision at both the membrane and the nucleus. This can only be achieved by attaining a fundamental understanding of the physical and chemical processes behind many aspects of the technology, including: kinetic understanding of fluorophores, theoretical fitting of impulse response functions and detailed knowledge of image reconstruction algorithms .
Selected Publications
For an up-to-date publications list, please see our Google scholar page.
Publications
- ‹ previous
- Page 2

