
Professor of Chemical and Molecular Biology
Protein Folding and Assembly
Our research focuses on different aspects of how proteins fold and how large protein complexes assemble. Projects cover diverse areas including biophysics, molecular biology (including protein engineering) and chemical biology. Current projects include:
How does a knotted protein fold?
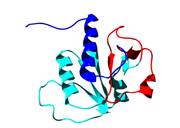
Recently, a remarkable new class of proteins have been discovered which have deep topological knots formed by the polypeptide backbone. These structures represent a new challenge for the protein folding community - not only has the protein to fold but in doing so it must also knot. We are studying the folding pathways of several knotted proteins, including YibK shown on the right, using a combination of experimental and computational techniques.
How do molecular chaperones work to assembly large molecular complexes?
Heat shock protein 90 (Hsp90) is a highly abundant and important protein in our cells which is the target of a new class of anti-tumour agents. It plays a key role in the assembly of a number of cellular complexes which are critical in cellular signal transduction pathways. We are combining a large number of biophysical techniques to understand the mode of action of Hsp90.
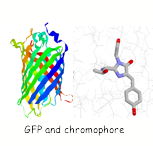
Single molecule studies on the folding of a large β-barrel protein.
The green fluorescent protein (GFP) first isolated from jellyfish is a protein with unique spectroscopic properties. A cyclisation and oxidation of the polypeptide backbone results in the formation of a chromophore which is highly fluorescent. In collaboration with the Klenerman group are using these special features of GFP to study the folding of this protein at a single molecule level.
Selected Publications
- Mallam, A.L., Rogers, J.M. and Jackson, S.E. (2010) Proc. Natl. Acad. Sci. 107, 8189-8194. Experimental detection of knotted conformations in denatured proteins
- Hsu, D., Blaser, G., Behrens, C., Cabrita, LD., Dobson, C.M. and Jackson, S.E. (2010) J. Biol. Chem. 285, 4859-4869. Folding Study of Venus Reveals a Strong Ion Dependence of its Yellow Fluorescence under Mildly Acidic Conditions
- Onuoha, S.C., Coulstock, E.C., Grossmann, J.G. and Jackson, S.E. (2008) J. Mol. Biol. 379, 732-744. Structural studies on the co-chaperone Hop and its complexes with Hsp90
- Mallam, A.L., Onuoha, S.C., Grossmann, J.G. and Jackson, S.E. (2008) Molecular Cell 30, 642-648. Knotted fusion proteins reveal unexpected possibilities in protein folding
- Orte, A., Craggs, T.D., White, S. Jackson, S.E. and Klenerman, D. (2008) J. Am.Chem.Soc. 130, 7898-7907. Evidence of an intermediate and parallel pathways in protein unfolding using single-molecule fluorescence
Watch Professor Jackson discuss her research
Take a tour of the Jackson Lab
Publications
- <
- 10 of 13
