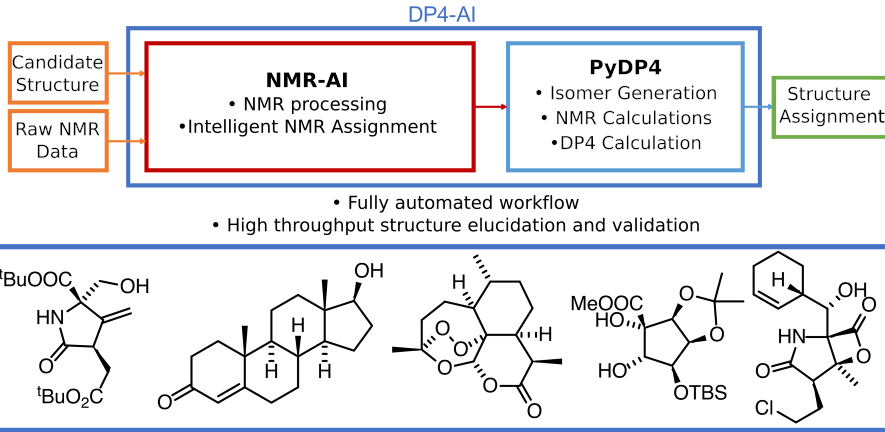
Every organic chemist faces the problem of structure elucidation, or the process of determining the chemical structure of a compound, often using NMR analysis.
Now Research Fellow Dr Kris Ermanis and PhD student Alex Howarth have developed a tool which, starting with just candidate structures and NMR data, can do the full calculation for a molecule in about 60 seconds, compared to up to eight hours if done manually.
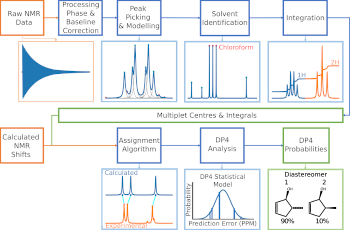
Building on an open source program called DP4, the team have developed DP4-AI, which “affords fully automated resolution of structural uncertainty, saving time interpreting NMR spectra, and giving confidence in the analysis,” says Professor Jonathan Goodman, who led the research.
Ermanis says in Nature blog Behind the Paper: “Five years ago when I joined Jonathan’s group, calculating DP4 probability involved lots of spreadsheets and the time-consuming task of managing computer time effectively.”
Ermanis decided that automating the process would be difficult but would ultimately lead to a quicker and more error-free process. In a vast understatement, he says: “This turned out to be much more difficult than I anticipated.”
Luckily, Ermanis was joined by postgraduate student Alex Howarth, who says in the blog: “I had the right combination of bravery and naivety to agree to take on the project.”
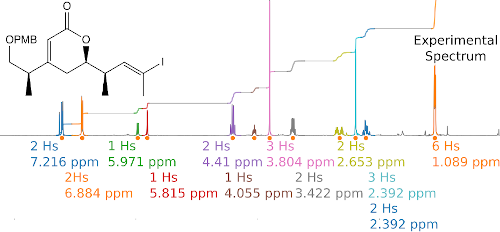
Together they worked hard to proceed from proof-of-concept to a robust process. They first found that it was difficult to obtain raw NMR FID data for testing, although this was eventually provided by several sympathetic synthetic groups.
Goodman notes that it was a particular challenge for the team to obtain a set of molecules with corresponding raw NMR data that was publically available. In an article in Chemistry World, Goodman observes: “Although many groups keep raw NMR data, the required labels and corresponding structures are often scribbled in dusty lab books that have been confined to a shelf. Even if the data is accessible, its meaning may quickly be lost. This will affect the reliability and how repeatable organic chemistry is.”
 Howarth and Ermanis emphasise that the chemical research community should think carefully about how this raw NMR data should be made more discoverable and accessible.
Howarth and Ermanis emphasise that the chemical research community should think carefully about how this raw NMR data should be made more discoverable and accessible.
Another challenge was to get the computer algorithms to think more like humans.
“Often, while staring at a spectrum the computer had very obviously assigned incorrectly, we would ask ourselves, ‘what was our thought process in determining that this assignment is wrong?’, which we would encode as probabilities, matrices and scores,” says Ermanis in the blog.
Five years on from Kris’s initial decision, DP4-AI is sure to be adopted as a popular approach to structural elucidation. “Structure elucidation from raw NMR data now works just as well as with manually interpreted NMR spectra,” says Kris. “A DP4 probability calculation that used to take about a day of human time, now takes just one minute!”


Alex Howarth (left) and Kristaps Ermanis: "The right combination of bravery and naivety"
“We hope DP4-AI and its components will be useful for integration into other automated NMR analysis workflows,” say the two researchers in their blog. “We are keen to develop this idea further in various directions, but we are just as excited to see what other people will do with it!”
DP4-AI Automated NMR Data Analysis: Straight from Spectrometer to Structure, A. Howarth, K. Ermanis and J. M. Goodman, Chemical Science 2020

