
We are interested in molecular recognition, aiming to uncover and exploit the rules governing non-covalent interactions. Hydrophobic, π–π, donor–acceptor, metal– ligand and hydrogen bonding interactions are used to create new supramolecular systems that expand our understanding of molecular behaviour and may have useful recognition, catalytic or photophysical properties. In particular in the past few years we have developed the concept of dynamic combinatorial chemistry as a new approach for discovering entirely unexpected structures and assemblies. Over the years our building blocks have included peptides, metalloporphyrins, steroids and simple aromatics, and our products have included macrocycles, rotaxanes, catenanes, molecular knots and supramolecular nanotubes. Very recently, while investigating dynamic chemistry in the solid state using ball mill grinding, we have discovered solvent and surface effects on polymorph stability in nanocrystals
Please note that I am not taking any new students or postdocs into my research group.
Selected Publications
Evolution of dynamic combinatorial chemistry, Accounts Chem. Res., (2012), 45, 2211.
Discovery of an organic trefoil knot, Science, (2012), 338, 783.
Templated dynamic synthesis of a [3]Catenane, Angew. Chemie Intl. Edn., (2012), 51, 1443.
Thermodynamics of supramolecular naphthalenediimide nanotubes, J. Am. Chem. Soc., (2012), 134, 566.
Discovery of linear receptors for multiple dihydrogen phosphate ions using dynamic combinatorial chemistry, J. Am. Chem. Soc., (2011), 133, 3804.
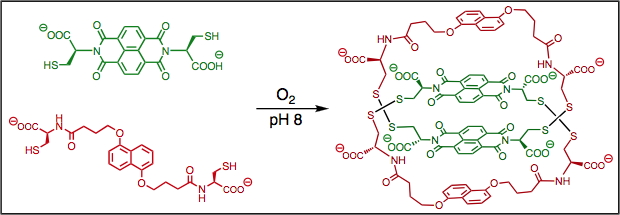
Formation pathways of Donor-Acceptor catenanes in aqueous dynamic combinatorial libraries, J. Am. Chem. Soc., (2011), 133, 3198.
Solid-state dynamic combinatorial chemistry, Chem. Sci., (2011), 2, 696.
An unexpected receptor for C70, Angew. Chemie Intl. Edn., (2008), 47, 2689.
Publications
- ‹ previous
- Page 11

