
Royal Society GSK Research Professor
Watching single molecules in action
We are physical scientists interested in developing and applying a range of new quantitative biophysical methods, based on single molecule fluorescence and scanning probe microscopy, to important problems in biology, which have not been addressed to date due to the lack of suitable tools and hence directly image important and disease relevant biological processes at the level of single molecules. While our experiments range from experiments in test-tubes to imaging living cells, no previous biological background is required for our research since we work closely with a range of biological and clinical collaborators. We are also part of the Cambridge Dementia Research centre. The best example of the success of this approach was the development of next generation DNA sequencing see the videos below:
https://www.youtube.com/watch?v=gAbFRUYlkZM
https://www.youtube.com/watch?v=FgGO2QtudAM
https://www.youtube.com/watch?v=T4RNbR4wZWw
Single molecule fluorescence
By studying molecules one at time, using fluorescence, specific complexes in a mixture can be identified and analysed without the need for any separation. With our collaborators we are exploiting single molecule fluorescence spectroscopy to study a range of biologically important molecules and processes. At present our main projects are:
- Imaging the early molecular events that lead to the triggering of T-cells, a process that underpins the adaptive immune response and is remarkably sensitive and selective.
- Imaging the early molecular events that lead to the triggering of Toll-like receptors, a key process in the innate immune response which plays an important role in causing inflammation in neurodegenerative diseases such as Alzheimer’s disease.
- Imaging and characterising the protein aggregates formed in the test-tube during aggregation reactions of disease-associated proteins and those present in human samples such as cerebral spinal fluid and brain tissue. We want to determine the aggregates responsible for causing neurodegenerative diseases such as Alzheimer’s and Parkinson’s disease in humans and the mechanisms by which they damage cells and spread through the brain.
- Determining the structure and organisation of DNA in the nucleus of cells and how this is regulated and changes during cell differentiation.
- Imaging aggregates of P53 and determining the role of these aggregates in the development and spreading of cancer.
- Developing and improving our imaging methods. At present our focus is developing methods to image protein aggregates at 20 nm spatial resolution and determine their composition and structure.
Scanning nanopipette
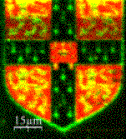 In collaboration with Professor Korchev at Imperial College we have developed a method based on a scanning nanopipette that allows robust, high resolution, non- contact imaging of living cells, down to the level of individual protein complexes. It can also be used to probe function by performing nanoscale assays such as locally deliver controlled amounts of reagents or performing single ion channel recording. The figure shows the University of Cambridge crest written in fluorescent DNA. We are using this method to watch the details of biological process taking place on the surface of living cells and to directly observe protein aggregates damage neuronal cells.
In collaboration with Professor Korchev at Imperial College we have developed a method based on a scanning nanopipette that allows robust, high resolution, non- contact imaging of living cells, down to the level of individual protein complexes. It can also be used to probe function by performing nanoscale assays such as locally deliver controlled amounts of reagents or performing single ion channel recording. The figure shows the University of Cambridge crest written in fluorescent DNA. We are using this method to watch the details of biological process taking place on the surface of living cells and to directly observe protein aggregates damage neuronal cells.
For a recent review of our work on neurodegenerative disease see: Imaging individual protein aggregates to follow aggregation and determine the role of aggregates in neurodegenerative disease. Biochimica et Biophysica Acta - Proteins and Proteomics 1867,870 (2019).
Two recent talks are also available on Youtube which provide an overview of the work in the group : https://www.youtube.com/watch?v=S6Dxzajz5iM and https://www.youtube.com/embed/bUE2uG3crBE
Watch Professor Klenerman discussing his research
Take a tour of the Klenerman Lab
Selected Publications
Super-resolution imaging unveils the self-replication of tau aggregates upon seeding. Cell Reports (2023) 42, 112725
Antigen discrimination by T cells relies on size-constrained microvillar contact. Nature Communications (2023) 14, 1611
Small soluble a-synuclein aggregates are the toxic species in Parkinson’s disease. Nature Communications (2022) 13, 5512
Structure-specific amyloid precipitation in biofluids Nature chemistry (2022) 14, 1045
In vivo rate-determining steps of tau seed accumulation in Alzheimer’s disease. Sci Adv (2021) 7, eabh1448
Different soluble aggregates of Abeta 42 can give rise to cellular toxicity through different mechanisms. Nat Communications (2019) 10,1541 .A
Direct Observation of the Interconversion of Normal and Toxic Forms of alpha-Synuclein. Cell (2012). 149, 1048-1059
Publications
- ‹ previous
- Page 6

