We can undertake collaborative work, please contact a member of the Mass Spec team to discuss.
Protein-Protein interactions
We studied the molecular mechanism by which the yeast Ctf4 protein links the Cdc45-MCM-GINS(CMG) DNA helicase to DNA polymerase α (Pol α) within the replisome. Native mass spectrometry, X-ray crystallography and electron microscopy showed that Ctf4 self associates as a trimer. By native mass spectrometry we also demonstrated that one Ctf4 trimer can support binding of up to three partner proteins, including the simultaneous association with both Pol α and GINS. Ctf4CTD (C-terminal domain) in the presence of the Ctf4-binding sequences of Pol α and Sld5 showed reconstitution of complexes with 1:1, 1:2 and 1:3 Ctf4-to-peptide stoichiometries. Native mass spectrometry analysis of the Ctf4CTD trimer in the presence of peptides of Pol α and Sld5 is shown in the spectra below.

A Ctf4 trimer couples the CMG helicase to DNA polymerase α in the eukaryotic replisome , Authors: Aline C. Simon, Jin C. Zhou, Rajika L. Perera, Frederick van Deursen, Cecile Evrin, Marina E. Ivanova, Mairi L. Kilkenny, Ludovic Renault, Svend Kjaer, Dijana Matak-Vinković, Karim Labib, Alessandro Costa & Luca Pellegrini (Nature) 2014.
Proteins and Protein Complexes
Native mass spectrometry is ideal tool for addressing questions about stoichiometry of protein complexes and the existence of subcomplexes in vivo and in vitro. We studied subcomplexes of the Polycomb Repressive Complex 2 (PRC2), such is heterodimer EZH2-EED. PRC2 complex interacts with ~20 % of long noncoding RNAs (lncRNAs) which have significant role in the regulation of gene expression. One of the most well-studied PRC2 interacting lncRNAs is HOX Transcript Antisense RNA (HOTAIR). EZH2-EED is the minimal HOTAIR Interacting unit of PRC2. Nano ESI mass spectra for intact EZH2-EED, showing presence of discrete oligomeric EZH2-EED complexes. The EED subunit alone did not show any oligomerization, further indicating oligomerization is specific to the EZH2-EED heterodimer. Our data show that oligomerization is intrinsic to EZH2-EED, and is likely to be a functional property of the heterodimer, rather than the result of nonspecific aggregation.
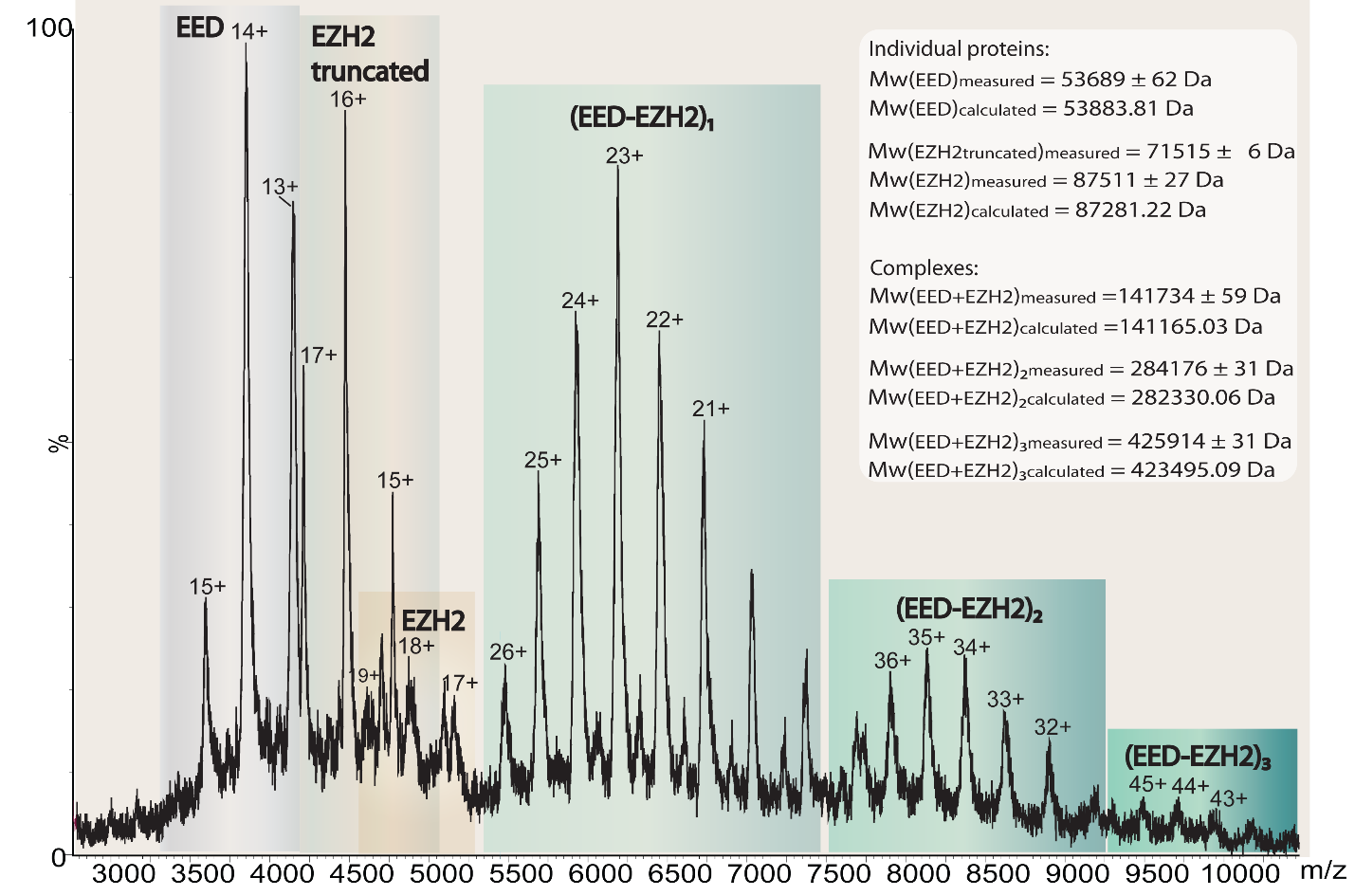
Binding Interactions between Long Noncoding RNA HOTAIR and PRC2 Proteins, Authors: Liang Wu, Pierre Murat, Dijana Matak-Vinkovic, Adele Murrell, Shankar Balasubramanian (Biochemistry) 2013.
Protein-Ligand Interactions
Native mass spectrometry is used for studying cytochrome P450s enzymes in interaction with small molecules. The molecular basis of the P450–ligand interactions, as well as the relationship between the structure and binding selectivity and affinity could facilitate the design of novel therapeutic agents. In our study P450 isoform CYP121 from human pathogen Mycobacterium tuberculosis (Mtb), which has been shown to be essential for Mtb viability in vitro, was analysed in interaction with inhibitors azoles, with the natural substrate cYY, and with novel compounds. Isolated unbound CYP121 is predominantly dimeric protein while in the presence of some inhibitors changes the oligomerisation state from dimeric to monomeric.
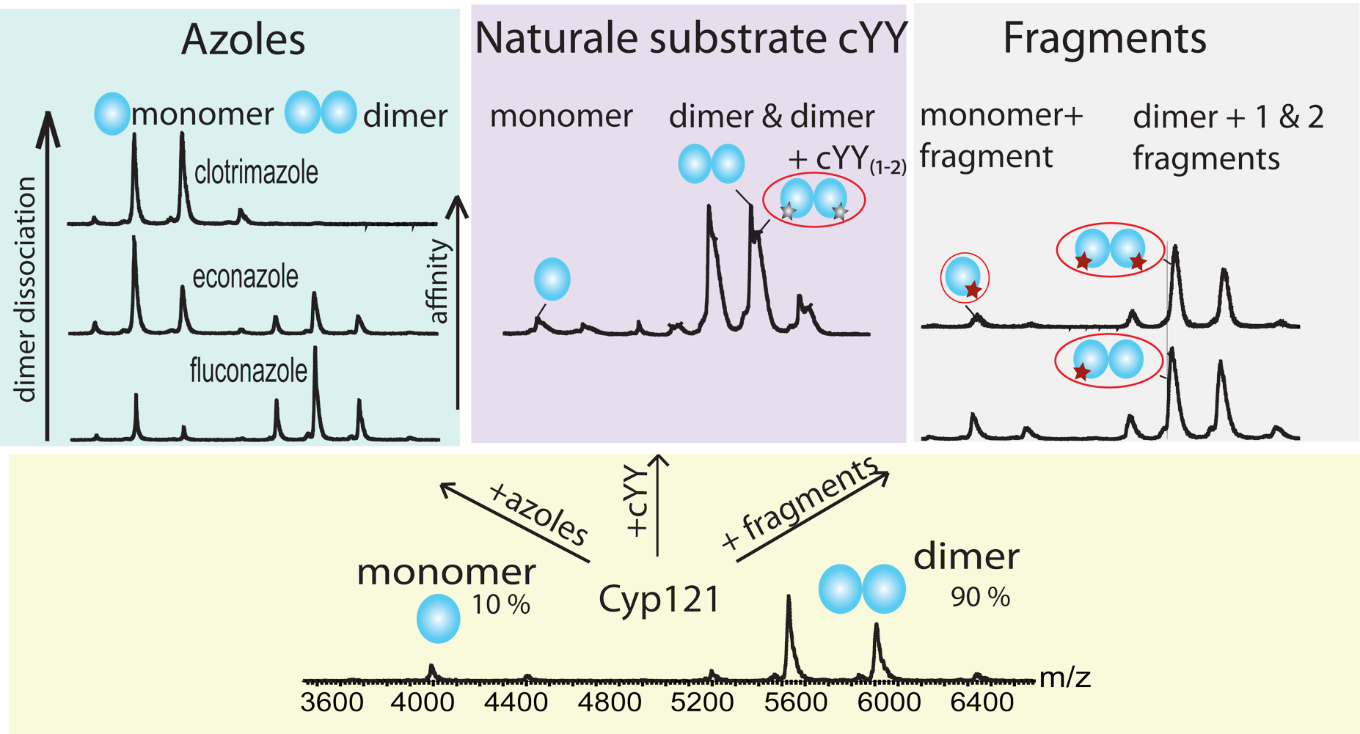
Nanoelectrospray Ionization Mass Spectrometric Study of Mycobacterium tuberculosis CYP121
Authors: Katie M. Duffell, Sean A. Hudson, Kirsty J. McLean, Andrew W. Munro, Chris Abell, Dijana Matak-Vinkovic (Analytical Chemistry) 2013.
Ion-mobility mass spectrometry
Ion mobility separation coupled to mass spectrometry (IM-MS) is used for the analysis of intact protein complexes. In IM-MS ions are separated based on their drift time, which depends on their cross section. Using IM-MS data the collision cross section for the ASB9−EloBC−Cul5NTD quaternary complex was determined.
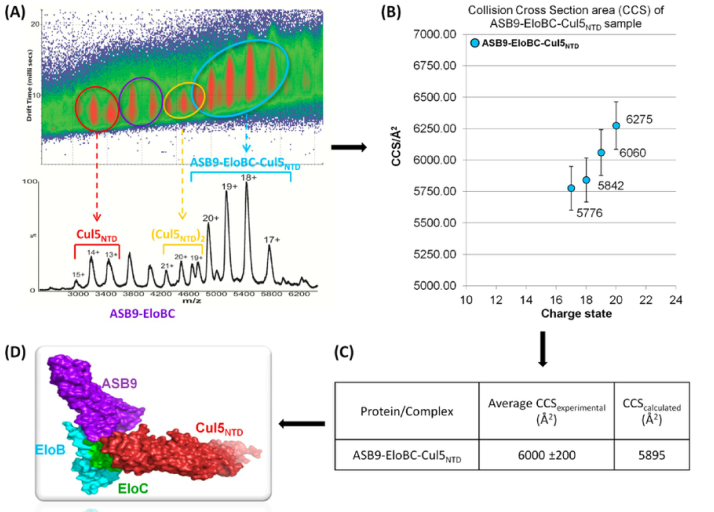
Multimeric Complexes among Ankyrin-Repeat and SOCS-box Protein 9 (ASB9), ElonginBC, and Cullin 5: Insights into the Structure and Assembly of ECS-type Cullin-RING E3 Ubiquitin Ligases
Authors: Jemima C. Thomas, Dijana Matak-Vinkovic, Inge Van Molle, Alessio Ciulli (Biochemistry) 2013.
