Inside cells, it has been shown that there are regions where the local concentration of some molecules is very considerably higher than elsewhere. Such membraneless compartments are sometimes thought to result from compositional (‘liquid–liquid’) phase separation of proteins and nucleic acids, which is a physical process similar to how oil and water mixtures separate into oil- and water-rich regions. Such a process would be energetically facile for cells, and so it is thought that such processes provide a mechanism for the spatiotemporal organisation of the cell nucleus.
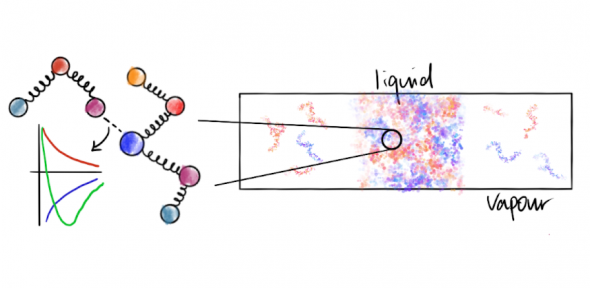
Because phase separation is a collective phenomenon, meaning that it results from the overall interactions of very many molecules, rather than the properties of the molecules in isolation, molecular modelling and simulations have become especially useful as powerful tools that can bridge physicochemical features of individual biomolecules to their modulation of the associated phase behaviour. However, since the number of atoms involved in such systems is extremely large, in order to be tractable, computer simulations must generally resort to coarse-graining, where many atoms (for example, an entire amino-acid residue) are represented as a single particle, and the solvent is treated only implicitly. Such an approach can give us qualitative insight into the fundamental processes governing biological phase separation. In order to gain more quantitative insight, we have joined forces with the Collepardo group and together we have designed a simple residue-level coarse-grained potential (‘Mpipi’) parameterised from a combination of atomistic simulations, bioinformatics data and experimental observations, and we have shown that it performs well in reproducing a range of experimental phase diagrams of intrinsically disordered protein systems.

Simulation of phase separation into three phases (two condensed phases and a vapour) from a fully mixed initial state using the Mpipi potential.
