Background:
Despite the importance of recognising complexity, fundamental investigations of the folding of small proteins remain at the heart of protein folding studies. Certain well-understood model systems have been central in the development of new conceptual frameworks. They provide test-beds for new theories and methods. Our group was the first to systematically set out to study the folding of families of related proteins - and now this has become an important tool in many laboratories1.

The contrasting topologies of some of the folds being studied by our group.
Mechanisms and pathway:
Studies of the folding of related proteins suggest that topology, the nature of the secondary structure content and sequence variations all play a role in determining how a protein folds2. A recent theoretical study suggests that topology is likely to be the dominant factor in determining folding pathways in all-beta proteins where long-range interactions predominate3. However, in all-alpha proteins, dominated by local interactions, sequence changes are more likely to result in variable folding pathways within a family.
Oliveberg and co-workers have analysed the nucleation of folding in a number of different families4. They describe nucleation motifs as "foldons", and suggest that they correspond to the smallest elements with sufficient stabilising interactions to overcome their loss in chain entropy5. The nuclei of the topologically complex Greek-key proteins comprise four elements of secondary structure6. Our investigations of basic folding studies aim to address the relative importance of topological complexity, chain connectivity, and secondary structure content on the folding mechanisms of proteins.
Landscape roughness - general or rare?
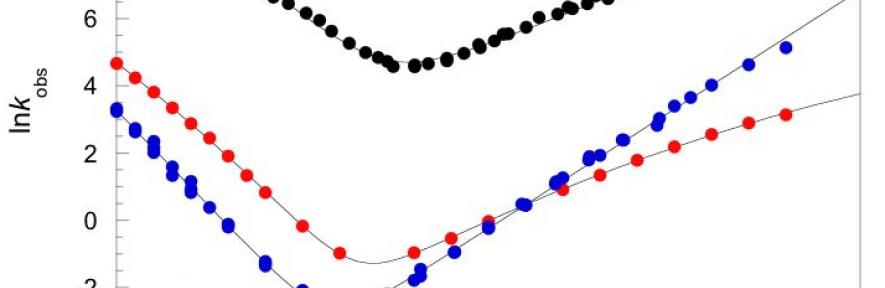
One of the principal observations from energy landscape theory is that, in general, evolution selects for smooth, frictionless landscapes, and that frustration (internal friction) would slow folding7. Our work, published in Nature in 2011, was the first direct demonstration of both the existence and relevance of significant landscape roughness in naturally occurring, slow-folding proteins (spectrin domains)8. This friction was observed by analysis of the viscosity dependence of the folding kinetics9. We proposed that introducing landscape roughness might be a mechanism whereby a protein might be able to be "long-lived" in vivo. We now intend to investigate the phenomenon of landscape roughness in more depth.
We wish to:
- understand the mechanism by which roughness is conferred.
- ask whether this is a common phenomenon that has not been detected because it has not been looked for.
- look for evolutionary evidence that roughness in proteins might be conserved as a protective mechanism against unfolding.
Current projects
- Spectrin Domains: investigating why some spectrin domains (e.g. R15) have low internal friction and fold rapidly, whereas almost identical sequences (e.g. R16 and R17) fold far more slowly with substantial internal friction.
- Death Domains: investigating whether secondary structure, chain connectivity or overall topology is most important for protein folding and stability.
- MATH domains: investigating whether circular permutants of β-sandwich proteins show altered folding mechanisms and increased susceptibility to aggregation.
- LysM domains: investigating whether small α+β proteins have transition states that are highly robust and resistant to mutation.
References
- Nickson, A. A. & Clarke, J. (2010). What lessons can be learned from studying the folding of homologous proteins? Methods 52, 38-50.
- Zarrine-Asfar, A., Larson, S. M. & Davidson, A. R. (2005). The family feud: Do proteins with similar structures fold via the same pathway? Curr. Opin. Struct. Biol. 15, 42-49.
- Cho, S. S., Levy, Y. & Wolynes, P. G. (2009). Quantitative criteria for native energetic heterogeneity influences in the prediction of protein folding kinetics. Proc. Natl. Acad. Sci. USA 106, 434-439.
- Shen, T. Y., Hofmann, C. P., Oliveberg, M. & Wolynes, P. G. (2005). Scanning malleable transition state ensembles: Comparing theory and experiment for folding protein U1A. Biochemistry 44, 6433-6439.
- Lindberg, M. O. & Oliveberg, M. (2007). Malleability of protein folding pathways: a simple reason for complex behaviour. Curr. Op. Struct. Biol. 17, 21-29.
- Lappalainen, I., Hurley, M. G. & Clarke, J. (2008). Plasticity within the obligatory folding nucleus of an immunoglobulin-like domain. J. Mol. Biol. 375, 547-559.
- Bryngleson, J. D. & Wolynes, P. G. (1989). Intermediates and barrier crossing in a random energy model (with applications to protein folding). J. Phys. Chem. 93, 6902-6915.
- Wensley, B. G., Batey, S., Bone, F. A. C., Chan, Z. M., Tumelty, N. R., Steward, A., Kwa, L. G., A., B. & Clarke, J. (2010). Experimental evidence for a frustrated energy landscape in a three-helix bundle protein family. Nature 463, 685-688.
- Cellmer, T., Henry, E. R., Hofrichter, J. & Eaton, W. A. (2008). Measuring internal friction of an ultrafast-folding protein. Proc. Natl Acad. Sci. USA 105, 18320-18325.
